Company
Platform
Insights
We profile tumor samples with our AI models. Next, we choose a large number of regions that are clinically meaningful and representative of overall sample biology to 'core'.
This lets us profile many samples in parallel underneath the exact same experimental conditions.
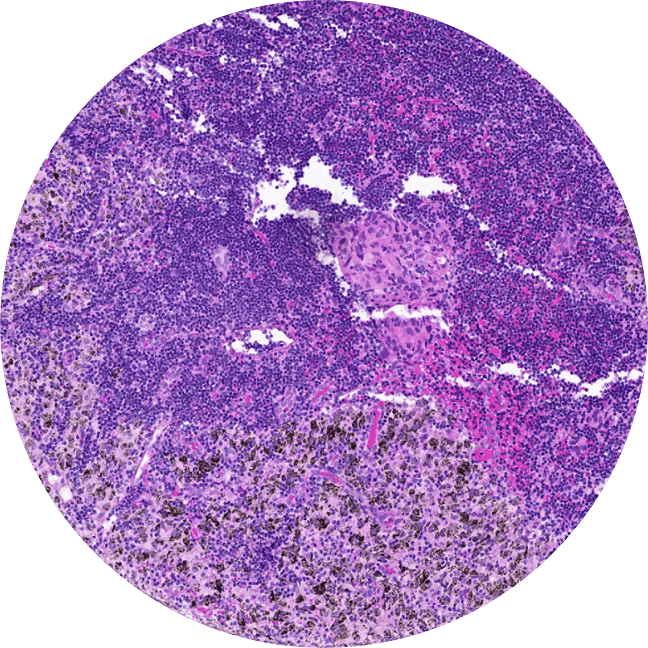
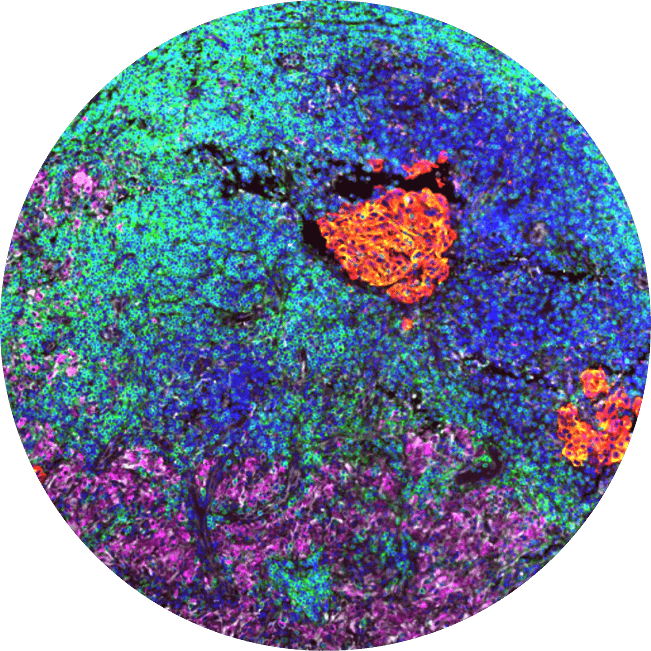
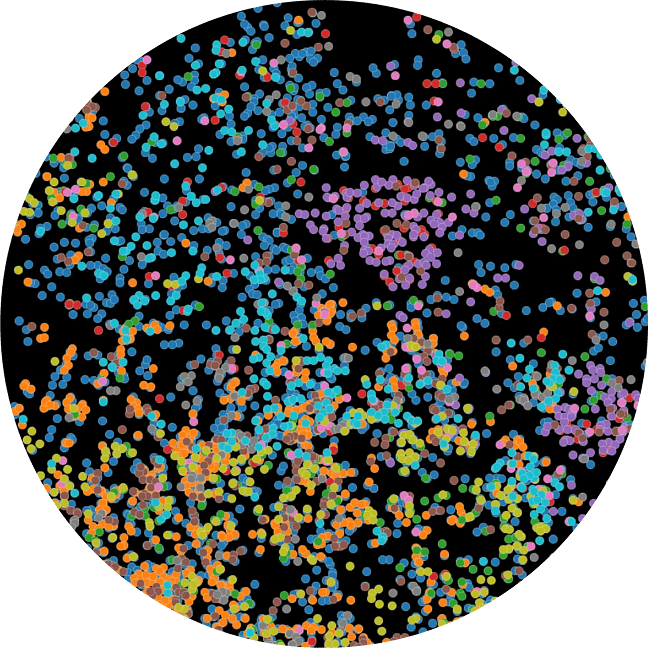
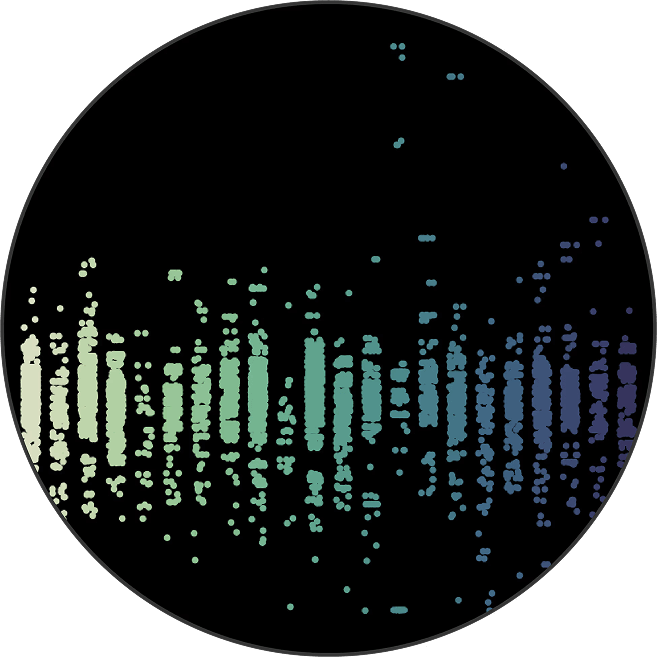
This is, by far, one of the largest of its kind by several orders of magnitude.