.png)
SAN FRANCISCO — Noetik, an AI-native biotech company leveraging self-supervised machine learning and high-throughput spatial data to develop next-generation cancer therapeutics, announced today the presentation of new research at the Society for Immunotherapy of Cancer (SITC) 2025 Annual Meeting in National Harbor, Maryland.
The poster titled “Virtual cell foundation model reveals genotype-specific fibroblast–macrophage crosstalk in the non-small cell lung cancer microenvironment”, highlighted new insights from OCTO-vc, Noetik’s spatial virtual cell foundation model. In addition, Eshed Margalit, Ph.D., Principal Machine Learning Scientist at Noetik, gave an invited talk titled “Simulating Virtual Cells in Human Tissue to Predict Treatment Outcomes: Use Cases Enabled by Multimodal Foundation Models” during Session II: AI in Synthetic Biology: From the Cell to the Organism.
The poster details how OCTO-vc simulates gene expression in virtual cells embedded within spatially resolved TMEs, allowing for in silico, cell-type–specific experiments. Yubin Xie, Ph.D., Senior Machine Learning Scientist at Noetik and presenter said “To uncover local cell–cell interactions between macrophages and tumor or fibroblast populations, Noetik designed virtual “clonal neighborhood” simulations, in which a virtual macrophage was surrounded by digital replicates of tumor or fibroblast cells sampled from NSCLC patients.” The team identified fibroblast- and tumor-mediated transcriptional programs associated with macrophage state thereby revealing previously uncharacterized spatial crosstalk patterns enriched in distinct NSCLC genotypes.
The invited talk describes Noetik's approach to using pre-trained foundation models of human cancer to predict therapeutic outcomes from clinical data. First, a foundation model is trained on a vast multimodal dataset collected in-house. The models are self-supervised and do not require data to be explicitly labelled. Then the pre-trained model is used to run millions of virtual cell simulations, each of which predicts how a virtual cell might behave when situated in different regions of a patient's tumor-immune microenvironment (TME). The simulations form a rich basis from which the space of human tumor biology is mapped. Finally, clinical data is used to identify which simulation outcomes are more common in responders versus non-responders. The features of responder-enriched simulation outcomes are studied to derive better biomarkers of treatment response. Critically, this approach is demonstrated to be viable for multi-modal models that learn to predict gene expression data from H&E images alone.
These findings demonstrate how virtual cell modeling can help decode spatial biology with implications for target discovery, biomarker development, and patient stratification in oncology.
The abstract will be published in the Proceedings of the Society for Immunotherapy of Cancer (SITC) 2025 Annual Meeting. SITC, the world’s leading organization dedicated to improving cancer patient outcomes through immunotherapy, is holding its Annual Meeting from November 4-9, 2025, at the Gaylord National Resort & Convention Center in National Harbor, Maryland.
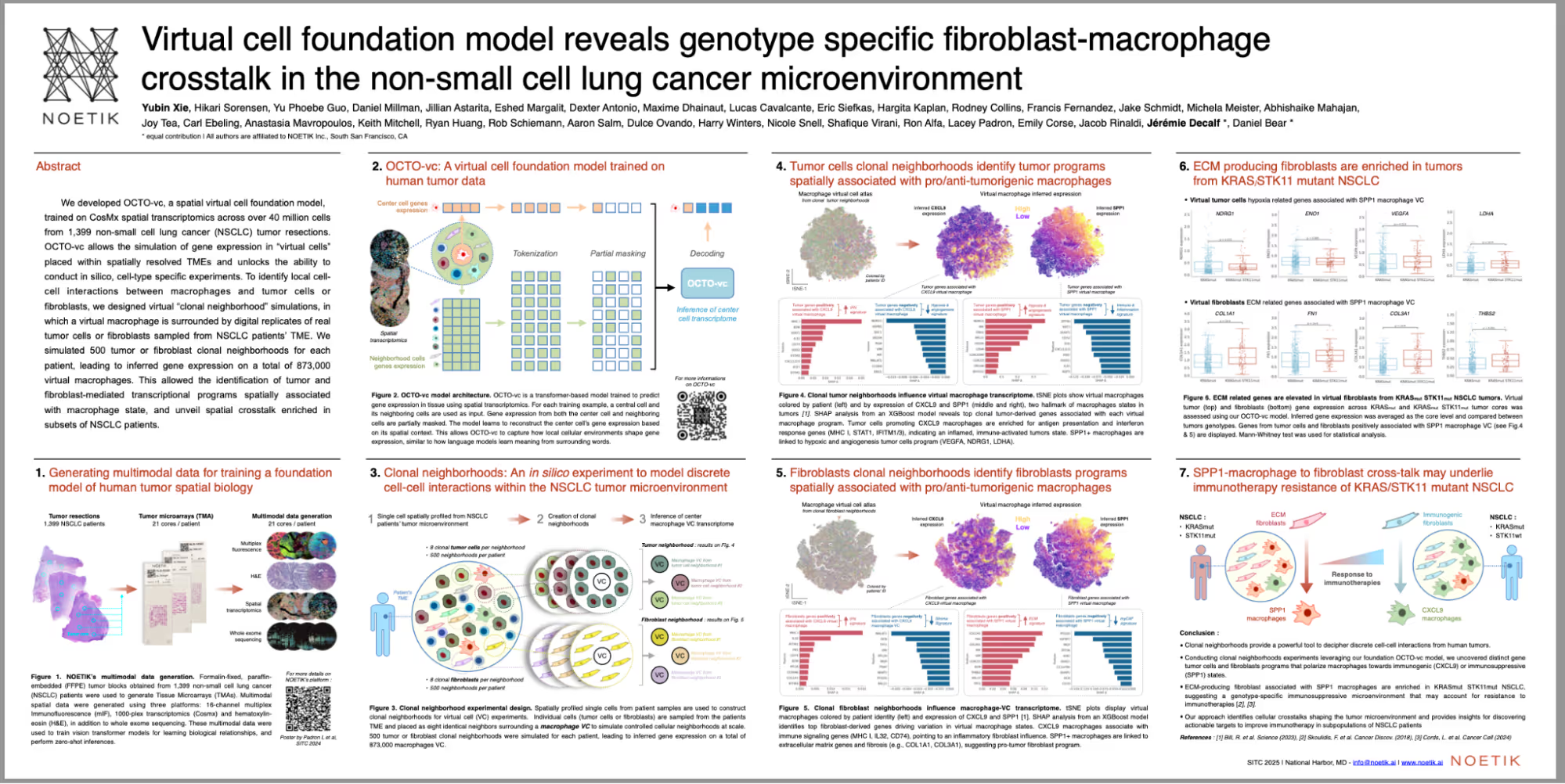
Virtual cell foundation model reveals genotype-specific fibroblast–macrophage crosstalk in the non-small cell lung cancer microenvironment
Abstract Number: 1130
Date: 11/8/25
